The PDF is free to download here.
Shared mutations in the human and chimpanzee β-globin pseudogenes is not evidence for a common ancestor
Evolutionists have suggested that the β-globin pseudogenes in humans and chimpanzees contain shared mutations and they have used this idea to conclude humans and chimpanzees share a common ancestor.1 Despite the recent discoveries that some pseudogenes actually have a function,2 the apparent β-globin pseudogenes and their so-called shared mutations are still being used as evidence for a common ancestor for humans and chimpanzees in the current literature.3 This conclusion is incompatible with biblical creation since the Bible says that humans and the ancestors of chimpanzees were created separately.
Human and chimpanzee β-globin gene clusters
Globin genes code for the predominate proteins in red blood cells—hemoglobin. It binds and transports oxygen from the lungs to cells throughout the body. Hemoglobin is needed because oxygen dissolves poorly in the blood plasma. Humans carry nine globin genes, which are all slightly different to each other. Five of these genes are clustered together in a region of DNA called the ‘β-globin gene cluster’, which is located on chromosome 11 in humans (figure 1). These genes are not all switched on at the same time but in stages, corresponding to their position on the chromosome and the different stages of human development.
The β-globin pseudogene in humans is located in the β-globin gene cluster, between the γ-A and the δ-globin genes (figure 1). There are two copies of the γ-globin genes, called γ-A and -G. The β-globin pseudogene shows higher similarity to the γ-globin gene, but is called the β-globin pseudogene because it was originally identified by comparing it to the β-globin gene of rabbits.4 Chimpanzees have the same globin genes in the same order, including the β-globin pseudogene.
Apparent genetic defects in the β-globin pseudogenes
Evolutionists hypothesized several point mutations and deletions in the β-globin pseudogenes of humans and chimpanzees and these differences render these regions incapable of being translated (figure 1). The start codon (a codon is a sequence of three adjacent nucleotides constituting the genetic code) of the human β-globin pseudogene has two apparent point mutations which prevents the protein-synthesizing machinery (ribosome) identifying it as a gene. A point mutation has been suggested in humans at codon 15 that signals the ribosome to prematurely terminate synthesis of the protein (premature stop codon). At codons 20 and 145 in humans it has been suggested that there are deletions of 1 bp. The first deletion messes up the code of the gene, resulting in six stop codons in the exons downstream, and the second scrambles a correct stop codon at the end of the pseudogene. These same hypothesized defects are found at the same position on the chimpanzee chromosome. In addition to the theoretical predictions, scientists have not detected any evidence using wet laboratory techniques that these pseudogenes in humans or chimpanzees are translated.4
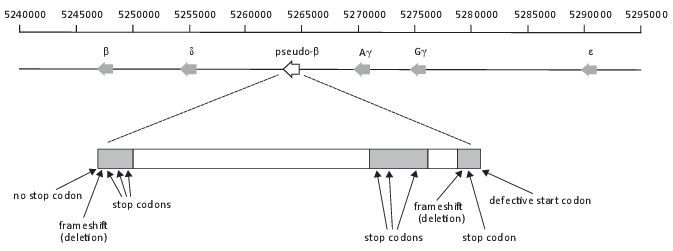
Figure 1. The organization of the
β-globin gene cluster in humans and an expanded view of the β-globin
pseudogene, showing the ‘defects’.
The first (horizontal) track displays the position on the chromosome (in
bp). The second track is a diagrammatic illustration of the
organization of β-globin gene cluster. Arrows indicate
the direction of the genes. The bottom bar is an expanded view of the
β-globin pseudogene. The white boxes are introns.
Interesting findings about the β-globin pseudogenes
There are three interesting findings about the β-globin pseudogenes from humans and chimpanzees. Firstly, when the pseudogenes from human and chimpanzee are aligned and analyzed for differences they are almost identical.1 The high level of similarity is not only seen in the exons of these pseudogenes, but also in the regions immediately upstream of the first exons (promoters and 5’ untranslated regions), between (introns), and immediately downstream of third exons (3’ untranslated regions). These regions have only 31 bp that are different. Secondly, there are putative promoters located upstream of these pseudogenes that are almost identical, with only 1 bp of the 15 bp TATA- and CAAT-like boxes that is different in humans and chimpanzees.1 The TATA- and CAAT-like boxes of the human β-globin pseudogene are located at nucleotides -25 and -81, respectively. The transcription initiation site is numbered as the +1 position and nucleotides upstream are numbered with negative integers. The human β-globin pseudogene TATA-like box has the sequence of TAAAAA and the CAAT-like box has the sequence GGTCAATAG. Normally, TATA and CAAT boxes are centered at nucleotides -25 and -80, respectively, with the consensus TATAAA and GGCCAATCT. Therefore, the TATA- and CAAT-like boxes of the human β-globin pseudogene are located at the correct positions and are only 1 and 3 bp, respectively, from the consensus sequence. Finally, the predicted intron junctions of the pseudogenes obey the ‘GU/AG RNA splicing rule’1 (‘RNA splicing’ is the modification of an RNA transcript, in which the transcript is cut at the intron junctions; the introns are then removed and the exons are spliced together).Evolutionists have shown little interest in these findings about the β-globin pseudogenes. This is because they have assumed that these pseudogenes are just more nonfunctional junk DNA. Evolutionists believe the DNA of more complex organisms, such as primates, must consist of mainly junk,5 so finding pseudogenes within the β-globin gene clusters is unsurprising to them. In addition to this, scientists may have detected that two exons of the β-globin pseudogene in humans are transcribed into non-coding RNA (ncRNA).6-8 However, this possible finding has been largely ignored by the scientific community, again because of the assumption that this region is a pseudogene. Some evolutionists prematurely dismiss this finding as merely noise in their wet laboratory results. They assume that many of the RNA transcripts detected in humans, primates and the other more complex organisms are because of the limitations of the current techniques and equipment used. Therefore, they have concluded these regions are not transcribed to RNA.9
Discussion
The fact that these β-globin pseudogenes in humans and chimpanzees are almost identical suggests that they either (1) have not been copied in each organism from generation to generation for the past five million years, (2) the entire regions, including the promoters, 5’ untranslated regions, introns and 3’ untranslated regions are functionally constrained and must remain almost identical, or (3) they have been transferred from chimpanzees to humans or vice versa, recently. The first suggestion is consistent with the Creation hypothesis. The second is unlikely, because Sanford has shown that natural selection cannot maintain the integrity of human DNA for so long.10 The third suggestion is also unlikely because there is no evidence in nature that large amounts of genetic material can be transferred between humans and chimpanzees. Consequently, the fact that high rates of mutations are not seen and that these regions are almost identical suggests they are young. Hence, this reduction in time for the existence of human and chimpanzee organisms makes it highly improbable they evolved from a common ancestor.
Based on the above evidence and our present understanding of genetics, the best hypothesis for these regions is that they function to remove the γ-globin RNA transcripts. As stated earlier in this article, subsets of globin genes are switched on and off according to the order on the chromosome, and their order correlates to the different stages of development. At the birth of a human infant, the γ-globin genes are significantly down-regulated and the β-globin gene is significantly up-regulated, with normal adult levels reached for each gene by the end of the first year of the infant’s life. The ncRNA transcripts from these pseudogenes may bind with ncRNA transcripts partners that are mirror images to the pseudogene transcripts, these transcripts are known as ‘antisense transcripts’. The pair of transcripts is then cut up, a process known as ‘dicing’, and the RNA fragments are used to direct degradation machinery to the γ-RNA transcripts. This process is known as ‘RNA induced RNA silencing’. Thus cellular levels of the γ-RNA transcripts are decreased. Although, the γ-globin genes may have their own antisense transcripts, a third antisense transcript would amplify the process of γ-globin RNA transcript removal.
- Since TATA- and CAAT-like boxes have been identified upstream of these regions, these regions may be transcriptionally active;
- the predicted intron junctions obey the GU/AG rule, therefore, suggesting that splicing would occur if these regions were transcribed;
- all three exons have at least one stop codon;
- these regions are similar to the γ-globin genes; and
- these regions are located between the γ-A and the β-globin genes.
Thus, evolutionists have somewhat prematurely concluded that these regions in humans and chimpanzees are genuine pseudogenes (i.e. former genes deactivated by mutations) and that there are shared mutations in these regions. Whatever the reason for these pseudogenes, there is a lack of evidence in these regions to suggest that humans and chimpanzees have a common ancestor.
Related Articles
Further Reading
References
- Chang, L.Y. and Slightom, J.L., Isolation and nucleotide sequence analysis of the β-type globin pseudogene from human, gorilla and chimpanzee, J. Mol. Biol. 180:767–784, 1984. Return to text.
- Sasidharan, R. and Gerstein, M., Genomics: protein fossils live on as RNA, Nature 453:729–731, 2008. Return to text.
- Miller, K.R., Only a Theory: Evolution and the Battle for America’s Soul, Penguin, New York, 2009. Return to text.
- Fritsch, E.F., Lawn, R.M. and Maniatis, T., Molecular cloning and characterization of the human β-like globin gene cluster, Cell 19:959–972, 1980. Return to text.
- ReMine, W.J., The Neutral Theory of Evolution; in: The Biotic Message: Evolution versus Message Theory, Saint Paul Science, Minnesota, MN, pp. 248–250, 1993. Return to text.
- Harrington, J J., Sherf, B., Rundlett, S. et al., Creation of genome-wide protein expression libraries using random activation of gene expression, Nat. Biotechnol. 19:440–445, 2001. Return to text.
- Kent, W.J., BLAT—the BLAST-like alignment tool, Genome Res. 12:56–64, 2002. Return to text.
- Hillier, L., Clark, N., Dubuque, T. et al., Unpublished work, Stanford University, Stanford, CA, 1995–1996. Return to text.
- Wang, J., Zhang, J., Zheng, H. et al., Mouse transcriptome: neutral evolution of ‘noncoding’ complementary DNAs, Nature 431:1–2, 2004. Return to text.
- Sanford, J.C., Genetic Entropy and the Mystery of the Genome, FMS Publications, Waterloo, NY, 2008. Return to text.
- Tam, O.H., Aravin, A.A., Stein, P. et al., Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes, Nature 453:534–538, 2008. Return to text.
